In phage-display, a gene encoding the surface protein of a bacteriophage is replaced with a gene encoding a novel protein. The technique can be used in drug discovery and research for the investigation of novel polypeptides.
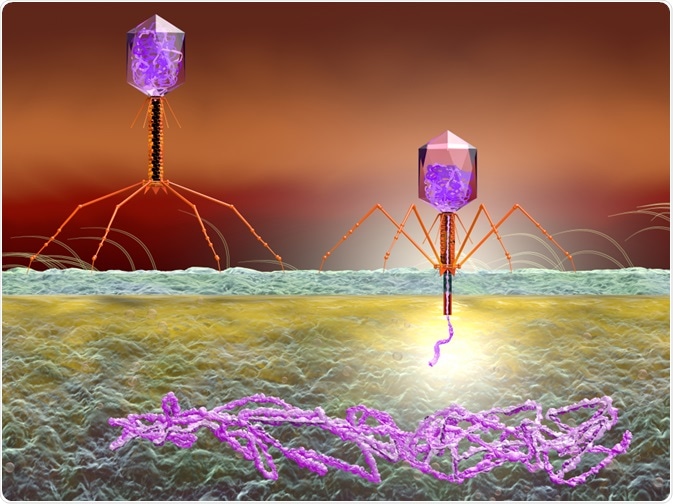 Image Credit: Axel_Kock / Shutterstock.com
Image Credit: Axel_Kock / Shutterstock.com
The discovery of phage display was spurred on by the discovery that filamentous phages can be synthetically engineered to exhibit amino acid sequences of foreign origin on the phage-coat proteins, without affecting the infectivity of the chosen phage.
Creating a phage library for antibody discovery
The potential of phage display is given by the links between modified coat proteins, and the genetics that encode for those proteins.
Each of the phage clones display a single sequence of peptides, however, a selection of phages end up displaying a vast variety of peptides. This library of peptides can be sequestered via a process called “affinity selection”, used to help in identifying specific phages, and which one is expressing a polypeptide with specific binding.
The process of affinity selection is built upon the immunological technique: panning, which includes elution and amplification. Once the phage/clone has been successfully isolated using the affinity selection method, DNA sequencing communicates the new, “foreign” DNA, with the displayed peptide binding to the relevant antibody.
The overall success of this new technology has been found to be due to a variety of factors. The insertion of a DNA sequence of foreign origin at a relevant site on the phage-coat protein leads to the presentation of that specific sequence on the phage’s surface.
Most notably, the phenotype of the peptide sequence displayed on the surface is linked to the sequence for encoding the ssDNA phage genome: the genotype. The link between genotype and phenotype has made it a possibility to choose a select peptide, followed by amplification via re-infecting E. coli bacteria and making them take in a vast amount of these peptides.
The analysis of these small volumes of the selected chemical (in this case, a specific peptide) are resolved simply, via the cost-effective method of virus replication, essentially using combinatory chemistry within a biological system.
How Phage Display is Used in Antibody Discovery
Using this phage display method, the development of vaccines is amongst a number of applications that have attracted interest in recent years. It allows researchers to pinpoint certain mimotopes, use phages directly as an immunogen, as well as study T-cell specificity.
Antigenic epitopes can be seen as a component of an antigen, which is involved in the interaction specific to the antigen-binding site of a T-cell receptor or antibody.
Obtaining detailed analyses of these epitopes is vital, both for the development of effective vaccines and tools for diagnosis of a variety of diseases, as well as the understanding of immune events. The characterization and identification of these epitopes can be an intricate process.
Epitopes are defined as ‘contact points’ of any biomolecular interaction within a biological system, unrestricted by interactions between proteins. The term ‘epitope’ is often used to discuss antigenic epitopes: such as the constituents of the antigen that is involved in binding to a specific antibody, such as a B-cell epitope, or T-cell epitope.
Even though multiple techniques have been discovered and further advanced in this field, there is still no simple, commonplace approach to this technique that can be used with all kinds of epitopes. Ever since its introduction over 10 years ago, phage display methods have had a significant impact within this field of research.
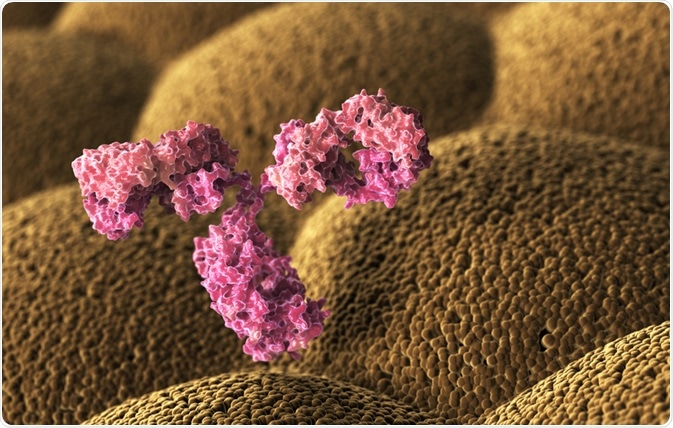 Image Credit: Tatiana Shepeleva / Shutterstock.com
Image Credit: Tatiana Shepeleva / Shutterstock.com
Take home message
The full potential of this technology in disease diagnosis and vaccine development has yet to be fully recognized, and the contribution of these methods to the study of epitopes goes way beyond being just another approach.
The considerable impact of phage display technology originates from its universality, simplicity, and the capability to identify new specific epitopes. Nonetheless, as is the case for most new emergent techniques, phage display has its difficulties.
Limitations posed by both host and phage biology could be more than has been expected, and it’s use in the investigation of T-cell epitopes is not as significant as it could be.
It is obvious that a clearer understanding of functional and structural characteristics of peptide-antibody interactions, as well as further clarification of some of the restrictions put in place by biological systems will one day soon advance the applicability and success fate of this already significantly powerful technology.
Further Reading