By Shelley Farrar, MSc, BSc
Green Fluorescent Protein (GFP) was first isolated from the jelly fish, Aequorea victoria. The protein consists of 238 amino acid residues that fluoresce green when exposed to UV light. The gene for GFP was successfully inserted into E.coli bacteria in 1994 and the Nobel Prize in Chemistry 2008 was jointly awarded to Osamu Shimomura, Martin Chalfie and Roger Y. Tsien for the discovery and development of GFPs.
They are commonly used as a reporter of expression in biology. Proteins can be tagged with GFP via genetic modification in order for a protein to be observed through fluorescence microscopy.
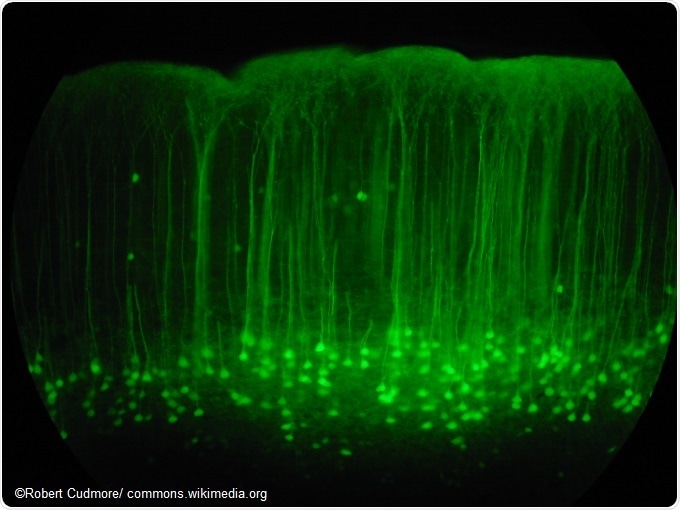
Somatosensory cortex mouse brain slice expressing GFP.
Fluorescence microscopy utilizes light to study specimens. A fluorescent chemical called a fluorophore is required that can absorb the light of specific wavelengths and then emit light of longer wavelengths. GFP-tagging is a way of preparing a sample for fluorescence microscopy by using the GFP as a fluorescent protein reporter.
This is done by cloning the GFP in frame with the target protein at either the N- or C-terminus of the amino acid chain. The use of GFP-tagging in fluorescence microscopy means that proteins within living cells can be visualized and studied.
The structure of GFP
The GFP that was first isolated from the jellyfish, Aequorea victoria, is known as wild-type or wtGFP and is the most widely used GFP. The structure of GFP includes the primary structure of a chain of 238 amino acids, and a secondary structure of hydrogen-bonded helices and pleated sheets and a tertiary barrel shaped structure capped with helices.
Within the center is the chromophore, an amino acid section responsible for the light emission. GFP is particularly useful for fluorescence microscopy as its formation means that the chromophore is encased in the barrel shaped structure, avoiding solvents and allowing the protein to fluoresce in many conditions. Furthermore, unlike other proteins that require enzymes for the complex folding necessary for the required protein shape, this occurs automatically in GFP and only additional oxidation of the chromophore is required for the protein to fluoresce.
The types of GFP
As wtGFP is able to absorb multiple wavelengths of light it is unsuitable for certain types of fluorescence microscopy. Other enhanced GFPs (eGFP) have been discovered, developed or created. One way of developing mutant GFPs is to allow colonies of bacteria with GFP expression to grow and wait for mutant variants of GFP to develop with time.
The majority of these mutations reduce fluorescence but some were found to increase brightness and photostability, as well as shifting the excitation peak to match the spectral characteristics of the commonly used light filter in fluorescence microscopy. Further mutations have increased the fluorescence yield meaning that there are now enough fluorescent proteins to cover nearly the whole visible spectrum.
Cell animation - fluorescence microscopy
The use of GFP-tagging in fluorescence microscopy including fluorescence resonance energy transfer (FRET)
Prior to GFP-tagging, the fluorescent molecules used in fluorescence microscopy were often phototoxic, as they harmed living cells through molecular changes when exposed to light. GFP-tagging allowed living cells to be illuminated without this type of destruction.
Early uses of GFP-tagging in fluorescence microscopy involved the protein being expressed in various structures to increase understanding of the differences in cellular morphology. The in-vivo imaging potential for fluorescence microscopy has been increased through the use of GFP-tagging, allowing for the observation of biological processes over time.
FRET is a technique that is used to observe real-time events within a cell. Two variants of GFP which respond to different wavelengths of light through absorption and emittance are bound to proteins. When the proteins are separated, light that excites one of the GFP variants will only cause a color to be observed from that protein. When the proteins interact and the same light is used, the two GFP variants become close enough together for there to be a transfer of energy between the excited donor fluorophore and the acceptor fluorophore.
This results in a color change to the emitted color of the second GFP. Through fluorescence microscopy, FRET can be used to quantify the distance between two fluorophores. Because FRET is distance dependant, this technique has been used to determine the structure of cells and the dynamics of protein interactions.
Sources:
- http://www.chm.bris.ac.uk/motm/GFP/GFPh.htm
- Yuste, R. 2005. Fluorescence microscopy today. Nature Methods, 2, pp. 902-904. https://www.nature.com/nmeth/journal/v2/n12/full/nmeth1205-902.html
- Chudakov, D. et al. 2005. Fluorescent proteins as a toolkit for in vivo imaging. Trends in Biotechnology, 23, pp. 605-613. https://www.ncbi.nlm.nih.gov/pubmed/16269193
- Hussain, S. 2012. An Introduction to Fluorescence Resonance Energy Transfer (FRET). Science Journal of Physics, ISSN:2276-6367. http://www.sjpub.org/sjp/abstract/sjp-268.html
Further Reading